

Illumina and PacBio/Oxford Nanopore data, legacy 454 and Sanger data
Illumina, Solexa, Sanger, 454, Ion Torrent, PacBio, Oxford Nanopore Protein-level assembler: assembles six-frame-translated sequencing reads into protein sequencesĪ suite of assemblers including de novo, metagenomic, ontology and taxonomic profiling uses a De Bruijn graph Illumina, ABI SOLiD, Roche 454, Ion Torrent, Solexa, Sanger Also de novo assembly and polishing of long read sequencing data from Oxford Nanopore and PacBio, including PacBio Hifi reads. Large genomes, exomes, transcriptomes, metagenomes, ESTs. Paired-end PCR-free reads (successor of ALLPATHS-LG)ĭNA sequence assembly with automatic end trimming & ambiguity correction. Parallel, paired-end sequence assembler designed for large genome assembly of short reads (genomic and transcriptomic), employ a Bloom filter to De Bruijn graph De Bruijn graph assemblers typically perform better on larger read sets than greedy algorithm assemblers (especially when they contain repeat regions).Ĭommonly used programs List of de-novo assemblers The assembler will then construct sequences based on the De Bruijn graph. Nodes that overlap by some amount (generally, k-1) are then connect by an edge. The k-mers are then used as nodes in the graph assembly. During the assembly of the De Bruijn graph, reads are broken into smaller fragments of a specified size, k. While both of these methods made progress towards better assemblies, the De Bruijn graph method has become the most popular in the age of next-generation sequencing. These methods represented an important step forward in sequence assembly, as they both use algorithms to reach a global optimum instead of a local optimum. String graph and De Bruijn graph method assemblers were introduced at a DIMACS workshop in 1994 by Waterman and Gene Myers. Graph method assemblers come in two varieties: string and De Bruijn. Some programs that used OLC algorithms featured filtration (to remove read pairs that will not overlap) and heuristic methods to increase speed of the analyses. These algorithms find overlap between all reads, use the overlap to determine a layout (or tiling) of the reads, and then produce a consensus sequence. Early de novo sequence assemblers, such as SEQAID (1984) and CAP (1992), used greedy algorithms, such as overlap-layout-consensus (OLC) algorithms. These algorithms typically do not work well for larger read sets, as they do not easily reach a global optimum in the assembly, and do not perform well on read sets that contain repeat regions. Greedy algorithm assemblers typically feature several steps: 1) pairwise distance calculation of reads, 2) clustering of reads with greatest overlap, 3) assembly of overlapping reads into larger contigs, and 4) repeat. Greedy algorithm assemblers are assemblers that find local optima in alignments of smaller reads.

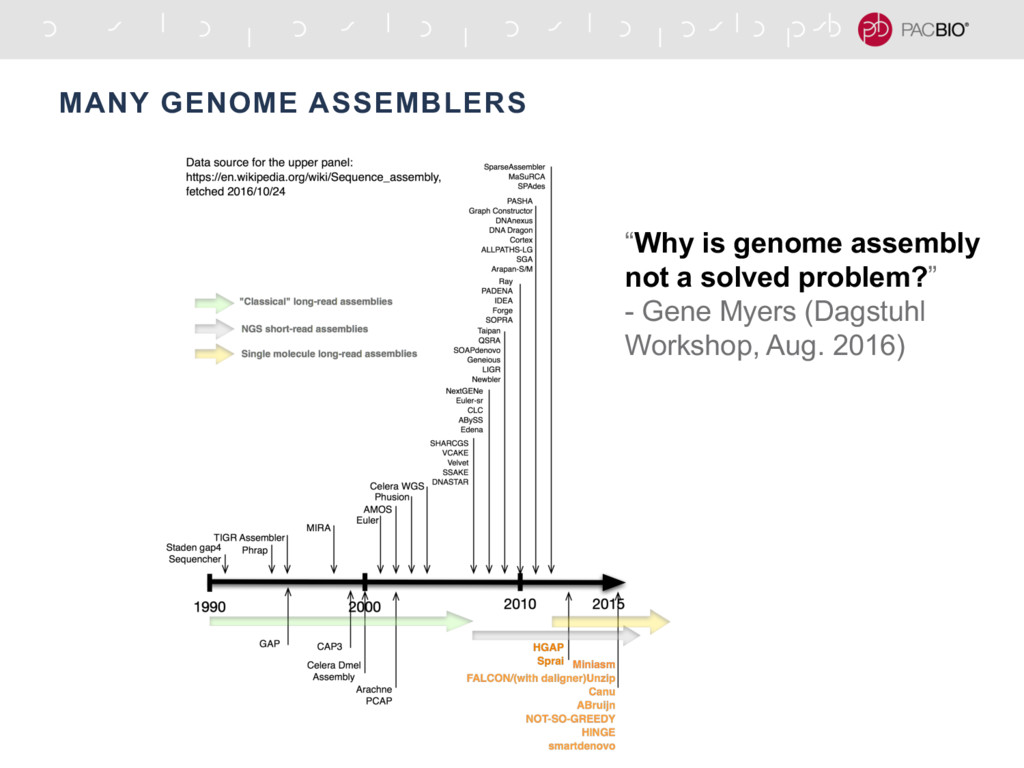
Different assemblers are tailored for particular needs, such as the assembly of (small) bacterial genomes, (large) eukaryotic genomes, or transcriptomes. There are two types of algorithms that are commonly utilized by these assemblers: greedy, which aim for local optima, and graph method algorithms, which aim for global optima. Two common types of de novo assemblers are greedy algorithm assemblers and De Bruijn graph assemblers. These are most commonly used in bioinformatic studies to assemble genomes or transcriptomes. De novo sequence assemblers are a type of program that assembles short nucleotide sequences into longer ones without the use of a reference genome.


 0 kommentar(er)
0 kommentar(er)
